| GO terms |
| P
| GO:0045893 |
Positive regulation of DNA-templated transcription |
|
| Transcription coactivator activity |
|
|
|
| Cysteine-type deubiquitinase activity |
|
|
|
| Positive regulation of DNA-templated transcription |
|
|
Interacting domains (7 domains)
Interacting motifs (1 motif)
HMM profile interface residues in EnY2 (7 interfaces)

| HMM prof. interface res. |
Binding partner(s) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
PDB entries that contain the EnY2 domain (19 structures)

| PDB |
Chains |
3did |
3fwb 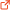
|
C |

|
3fwc 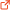
|
C, D, G, H,... |

|
3kik 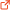
|
A, B, C, D |

|
3kjl 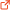
|
A, B, C, D |

|
3m99 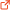
|
C |

|
3mhh 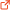
|
B |

|
3mhs 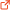
|
B |

|
4c31 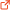
|
B, E |

|
4dhx 
|
B, C, E, F |

|
4fip 
|
B, F |

|
4fjc 
|
B, F |

|
4fk5 
|
B |

|
4mbe 
|
C, F |

|
4w4u 
|
B, F |

|
4wa6 
|
B, F |

|
4zux 
|
a, f, k, V |

|
6aqr 
|
B |

|
6t9l 
|
L |

|
6tbm 
|
P |

|